Research
- Research Topics
- Cell Biology and Tumor Biology
- Stem Cells and Cancer
- Inflammatory Stress in Stem Cells
- Experimental Hematology
- Molecular Embryology
- Signal Transduction and Growth Control
- Epigenetics
- Redox Regulation
- Vascular Oncology and Metastasis
- Clinical Neurobiology
- Molecular Neurogenetics
- Chaperones and Proteases
- Vascular Signaling and Cancer
- Molecular Neurobiology
- Mechanisms Regulating Gene Expression
- Molecular Biology of Centrosomes and Cilia
- Dermato-Oncology
- Pediatric Leukemia
- Tumour Metabolism and Microenvironment
- Personalized Medical Oncology
- Molecular Hematology - Oncology
- Cancer Progression and Metastasis
- Translational Surgical Oncology
- Neuronal Signaling and Morphogenesis
- Cell Signaling and Metabolism
- Cell Fate Engineering and Disease Modeling
- Cancer Drug Development
- Cell Morphogenesis and Signal Transduction
- Functional and Structural Genomics
- Molecular Genome Analysis
- Molecular Genetics
- Pediatric Neurooncology
- Cancer Genome Research
- Chromatin Networks
- Functional Genome Analysis
- Theoretical Systems Biology
- Neuroblastoma Genomics
- Signaling and Functional Genomics
- Signal Transduction in Cancer and Metabolism
- RNA Biology and Cancer
- Systems Biology of Signal Transduction
- Areas of Interest
- Advancement of clinical proteomics for systems medicine
- Bridging from the single cell to the cell population – Epo-induced cellular responses and erythroleukemia
- Deciphering tumor microenvironment interactions determining lung cancer development
- Mechanisms controlling the compensation of liver injury and towards model-based biomarkers for early detection of liver cancer
- Application of dynamic pathway modelling for personalized medicine
- Group Members
- Publications
- Open Positions
- Funding
- Teaching
- Areas of Interest
- Molecular thoracic Oncology
- Proteomics of Stem Cells and Cancer
- Computational Genomics and System Genetics
- Applied Functional Genomics
- Applied Bioinformatics
- Translational Medical Oncology
- Metabolic crosstalk in cancer
- Pediatric Glioma Research
- Cancer Epigenomics
- Translational Pediatric Sarcoma Research
- Artificial Intelligence in Oncology
- Mechanisms of Genomic Variation and Data Science
- Neuropathology
- Pediatric Oncology
- Neurooncology
- Somatic Evolution and Early Detection
- Translational Control and Metabolism
- Soft-Tissue Sarcoma
- Precision Sarcoma Research
- Brain Mosaicism and Tumorigenesis
- Mechanisms of Genome Control
- Translational Gastrointestinal Oncology and Preclinical Models
- Translational Lymphoma Research
- Mechanisms of Leukemogenesis
- Genome Instability in Tumors
- Developmental Origins of Pediatric Cancer
- Brain Tumor Translational Targets
- Translational Functional Cancer Genomics
- Regulatory Genomics and Cancer Evolution
- SPRINT
- Cancer Risk Factors and Prevention
- Cancer Epidemiology
- Biostatistics
- Clinical Epidemiology and Aging Research
- Health Economics
- Physical Activity, Prevention and Cancer
- Preventive Oncology
- Personalized Early Detection of Prostate Cancer
- Digital Biomarkers for Oncology
- Genomic Epidemiology
- Cancer Survivorship
- Immunology and Cancer
- Cellular Immunology
- Molecular Oncology of Gastrointestinal Tumors
- Immunoproteomics
- T Cell Metabolism
- Personalized Immunotherapy
- mRNA Cancer Immunotherapies
- Translational Immunotherapy
- B Cell Immunology
- Immune Diversity
- Structural Biology of Infection and Immunity
- Applied Tumor Immunity
- Neuroimmunology and Brain Tumor Immunology
- Adaptive Immunity and Lymphoma
- Dermal Oncoimmunology
- Immune Regulation in Cancer
- Systems Immunology and Single Cell Biology
- GMP & T Cell Therapy
- Immune Monitoring
- News
- Imaging and Radiooncology
- Radiology
- Research
- Computational Radiology Research Group
- Contrast Agents In Radiology Research Group
- Neuro-Oncologic Imaging Research Group
- Radiological Early Response Assessment Of Modern Cancer Therapies
- Imaging In Monoclonal Plasma Cell Disorders
- 7 Tesla MRI - Novel Imaging Biomarkers
- Functional Imaging
- Visualization And Forensic Imaging
- PET/MRI
- Dual- and Multienergy CT
- Radiomics Research Group
- Prostate Research Group
- Breast Imaging Research Group
- Bone marrow
- Musculoskeletal Imaging
- Microstructural Imaging Research Group
- Staff
- Patients
- Research
- Medical Physics in Radiology
- X-Ray Imaging and Computed Tomography
- Federated Information Systems
- Translational Molecular Imaging
- Medical Physics in Radiation Oncology
- Biomedical Physics in Radiation Oncology
- Intelligent Medical Systems
- Medical Image Computing
- Radiooncology - Radiobiology
- Smart Technologies for Tumor Therapy
- Radiation Oncology
- Molecular Radiooncology
- Nuclear Medicine
- Translational Radiation Oncology
- Molecular Biology of Systemic Radiotherapy
- Interactive Machine Learning
- Multiparametric methods for early detection of prostate cancer
- Molecular Mechanisms of Head and Neck Tumors
- Radiology
- Infection, Inflammation and Cancer
- Tumor Virology
- Viral Transformation Mechanisms
- Pathogenesis of Virus-Associated Tumors
- Immunotherapy and Immunoprevention
- Applied Tumor Biology
- Virotherapy
- Virus-associated Carcinogenesis
- Chronic Inflammation and Cancer
- Microbiome and Cancer
- Cell Plasticity and Epigenetic Remodeling
- Experimental Hepatology, Inflammation and Cancer
- Infections and Cancer Epidemiology
- Tumorvirus-specific Vaccination Strategies
- Mammalian Cell Cycle Control Mechanisms
- Molecular Therapy of Virus-Associated Cancers
- DNA Vectors
- Episomal-Persistent DNA in Cancer- and Chronic Diseases
- Cell Biology and Tumor Biology
- Research Groups A-Z
- Junior Research Groups
- Core Facilities
- Center for Preclinical Research
- Chemical Biology Core Facility
- Electron Microscopy
- Flow Cytometry
- Genomics and Proteomics
- Information Technology
- Library
- Kataloge -- Catalogues
- Zeitschriften - Journals
- E-Books - ebooks
- Datenbanken - Databases
- Dokument-Lieferung - Document Delivery
- Publikationsdatenbank - Publication database
- DKFZ Archiv - DKFZ Archive
- Open Access
- Science 2.0
- Ansprechpartner - Contact
- More Information - Service
- Anschrift - Address
- Antiquariat - Second Hand
- Aufstellungssystematik - Shelf Classification
- Ausleihe - Circulation
- Benutzerhinweise - Library Use
- Beschaffungsvorschläge - Desiderata
- Fakten und Zahlen - Facts and Numbers
- Kooperationen, Konsortien - Cooperations, Consortia
- Kopieren, Scannen - Copying, Scans
- Kurse, Führungen - Courses, Introductions
- DKFZ-Intern - internal only
- DEAL-Info
- Light Microscopy
- Omics IT and Data Management Core Facility
- Small Animal Imaging
- Metabolomics Core Technology Platform
- Data Science @ DKFZ
- INFORM
- Baden-Württemberg Cancer Registry
- Cooperations & Networks
- National Cooperations
- International Cooperations
- Cooperational Research Program with Israel: DKFZ - MOST in Cancer Research
- Program
- Members of the Program Committee
- Call
- Publication Database
- German-Israeli Cancer Research Schools
- Archive
- Heidelberg - Israel, Science and Culture
- Symposium 40 Years of German-Israeli Cooperation
- 35th Anniversary Symposium
- 34th Meeting of the DKFZ-MOST Program
- 40th Anniversary Publication
- 30th Anniversary Publication
- 20th Anniversary Publication
- Flyer - The Cancer Cooperation Program
- List Publications 1976-2004
- Highlight-Projects
- Cooperational Research Program with Israel: DKFZ - MOST in Cancer Research
- Cooperations with industrial companies
- DKFZ PostDoc Network
- Cross Program Topic RNA@DKFZ
- Cross Program Topic Epigenetics@dkfz
- Cross Program Topic Single Cell Sequencing
- WHO Collaborating Centers
- DKFZ Site Dresden
- Health + Life Science Alliance Heidelberg Mannheim
Anthropomorphic and zoomorphic phantoms with integrated tumor tissues
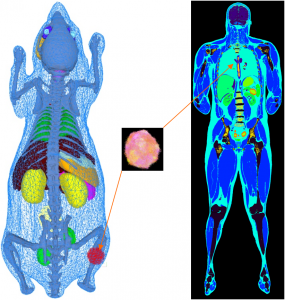
© dkfz.de
In our simulations packages such as Musiré, a single object description (phantom, atlas) can be used for the simulation of all imaging modalities. Whereas voxelized phantoms are generally employed for SPECT, PET, CT and MRI simulations, mesh-based representation of anatomical structures is preferred for the simulation of optical photons (BLI, FMI) since the imaged object's boundary contributes crucial information on the emitted photon flux which constitutes a decisive factor for image reconstruction or inverse light field mapping accuracy.
Our phantoms include (i) the three-dimensional (x, y, z) voxelized tissue/material atlas encoded as MetaIO (mhd) and (ii) the three-dimensional polygonal (triangle) mesh for tissue atlas geometry representing general anatomy (and objects) encoded in the Stanford triangle format (ply). Our programs are also compatible with three-dimensional point clouds encoded as ply. A multitude of meshes and point clouds can be encapsulated within a MeshLab project (mlp) container. From these, our frameworks automatically convert the object description into the appropriate format for every simulator. The mechanism of passing parameter files or scripts also allows for use of simulator-specific phantom representation models. Physical attributes such as the definition of materials, densities, source activities, dye concentrations, relaxation times and others are assigned to mhd or ply represented atlas regions by simple look-up table (dat) files. Point clouds are intended for representing (multi-lesion) heterogeneous tumour cell distributions such as generated by biologically derived models for tumour growth dynamics. Anatomical phantoms can be extended with arbitrarily placeable tumour inserts. These inserts can represent arbitrarily solid, multi-lesion, necrotic or heterogeneous entities with millions of cells per cubic centimetre of tissue. Depending on the simulated imaging modality, point clouds can be used at their native resolution. This is implemented for optical imaging where bioluminescence light distribution is sampled directly from the spatial tumour cell distribution. However, considering the size of tumour cells to be of the order of 10 μm to 50 μm it would be inefficient to use this degree of spatial resolution, e.g. in nuclear imaging simulation. Hence, point cloud tumour inserts are being down-sampled automatically for other modalities into the spatial resolution of the host phantom. By employing specific command-line options tumour cell density per voxel can be scaled into a source activity range or further physical material entities, such as T1 and T2 relaxation times.
(← back to research group home web page)
