Emmy Noether Research Group Epigenetic Machineries and Cancer
Indrabahadur Singh, PhD

Indrabahadur Singh, PhD
Emmy Noether Research Group Epigenetic Machineries and Cancer
Chronic Inflammation and Cancer (F180)
Deutsches Krebsforschungszentrum
Im Neuenheimer Feld 280
69120 Heidelberg
Tel: +49 6221 42 3894
E-Mail
Research
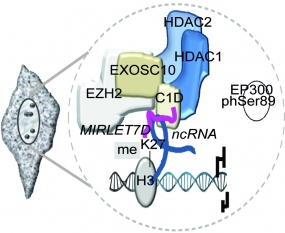
Figure: Novel evolutionarily conserved epigenetic machinery, named MiCEE complex. MiCEE = Mirlet7d + C1D + EXOSC10 + EZH2 (Singh I et al, Nat Genet 2018, Rubio K et al, Nat Commun 2019). Our aim is to understand altered epigenetic machineries in pathological conditions and to identify novel strategies for precision therapy.
© dkfz.de
Multiple cell-signaling pathways converge on chromatin to induce gene expression programs. The epigenetic machineries tightly regulate the chromatin structure in a manner that alters the availability of a gene’s promoter to transcription factors without altering the primary DNA sequence. Chromatin is the physiological template for DNA-dependent processes, including replication, recombination, repair and transcription. Accessibility to DNA within chromatin is a central factor that determines the DNA-dependent nuclear processes. Although altered chromatin organization has long been recognized as a feature of cancer, the molecular underpinnings of chromatin structure, epigenetic regulation, and their relationships to transcription are only beginning to be understood.
The Epigenetic machineries and Cancer group funded by the Emmy Noether Program of the German Research Foundation (DFG) has been established with the aim to understand the altered epigenetic machineries underlying non-alcoholic fatty liver disease (NAFLD), viral infections and cancer as well as translate this knowledge for personalized epigenetic therapy. This is achieved by systematic investigation of specific altered epigenetic machineries in NAFLD and cancer employing current state-of-the-art technologies of functional genomics, genome editing methods (CRISPR/CAS9), pre-clinical mouse models, cell culture and human patient samples.
We are currently pursuing the following objectives:
- Investigation of dysregulated chromatin remodeling factors in non-alcoholic fatty liver disease and liver cancer
- Mechanistic underpinnings of epigenetic machineries
- Identification of epigenetic signatures for spatial stratification of intratumoral heterogeneity.
These studies will identify molecular mechanism-guided therapeutic interventions for metabolic syndrome and cancer.
Selected publications:
Rubio K, Singh I# et. al (2019). Inactivation of nuclear histone deacetylases by EP300 disrupts the MiCEE complex in idiopathic pulmonary fibrosis. Nature Communications. 10(1):2229.
Singh I, Contreras A et. al (2018). MiCEE is a ncRNA-protein complex that mediates epigenetic silencing and nucleolar organization. Nature Genetics. 50(7):990-1001.
Mehta A*, Cordero J* et. al (2016). Non-invasive lung cancer diagnosis by detection of GATA6 and NKX2-1 isoforms in exhaled breath condensate. EMBO Molecular Medicine. 8(12):1380-1389.
Singh I, Ozturk N et. al (2015). High mobility group protein-mediated transcription requires DNA damage marker γ-H2AX. Cell Research. 25(7):837-50.
Patent:
Barreto G, Mehta A, Singh I, Szibor M, Savai R, Seeger W, Braun T, Günther A, Krüger M. ‘Specific isoforms of GATA6 and NKX2-1 as markers for the diagnosis and therapy of cancer and as targets for anti-cancer therapy.’ European patent PCT/EP2014/060489. Issued 2018.
