Division of Neuroblastoma Genomics
PD Dr. Frank Westermann
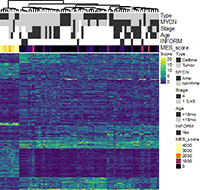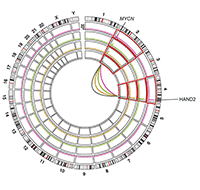
Figure 1: Clustering based on enhancer activity as defined by tumor-ChiP-seq.
Figure 2: Translocation event in neuroblastoma cells analyzed using 4C– enhancer hijacking.
© dkfz.de
Neuroblastoma (NB) is the most common pediatric single-entity solid cancer derived from primitive cells of the peripheral sympathetic nervous system. It is characterized by heterogeneous clinical phenotypes ranging from spontaneous regression to malignant progression despite intensive multimodal therapies. The presence of an active telomere maintenance mechanism is associated with aggressive growth and poor outcome in neuroblastoma, while low-risk tumors usually lack a telomere maintenance mechanism. Subsets of high-risk neuroblastomas elongate telomeres either by telomerase activation (as a result of amplified MYCN or rearranged TERT) or by alternative lengthening of telomeres (ALT).
Overall aim of our work is to integrate next-generation molecular diagnostics for a more precise patient risk stratification and to develop molecularly targeted therapies based on a better understanding of the molecular mechanisms underlying neuroblastoma tumorigenesis.
We use multi-omics high-throughput profiling with the specific aim to define genetic, epigenetic and metabolic alterations associated with distinct neuroblastoma subtypes. Furthermore, we aim at understanding the enhancer regulated core regulatory circuitries defining the epigenome and transcriptome of neuroblastoma cells and their role in oncogenesis, relapse and metastasis formation. Using an integrated analysis combining whole genome sequencing (WGS), RNA-sequencing, ChIP-sequencing and 4C/HiC chromatin context analyses, we compile a comprehensive characterization of enhancer translocations in neuroblastoma cells. With the help of single-cell sequencing technologies in combination with spatial transcriptomics of tumor cells in comparison to normal embryonal and fetal tissues during development, we investigate the developmental origins and evolutionary trajectories of neuroblastoma tumors. Our (epi)genetic work is complemented by the study of deregulated metabolic networks in neuroblastoma cells and how these can provide an Achilles heel for cancer therapy.
To disclose neuroblastoma-specific vulnerabilities associated with distinct genetic aberrations and to systematically identify specific targets for personalized medicine approaches, we use high-throughput genetic and compound screens. We have a long-standing expertise in preclinical target development and validation in close collaboration with clinical and industry partners (e.g., the development of proteolysis targeting chimeras (PROTACs) to specifically degrade unwanted proteins in cancer cells).
Combining our high-throughput genetic and molecular data with systems medicine approaches, we want to gain insight into the genetic evolution of neuroblastoma tumors and understand the underlying complex oncogenic networks.
NB super-enhancer visualization tool
NB developmental programs visualization tool

